Anna Syme
Click name ↑ to return to homepage
Research
Links to publications, presentations, and training resources developed.
Bioinformatician, Melbourne Bioinformatics and the Australian BioCommons
The University of Melbourne, October 2020 to present
Role: Workforce transition and user-upskilling for the Australian BioCommons Bring-Your-Own-Data project. Project link: https://www.biocommons.org.au/byod-expansion. Some research updates:
November 2021
-
I have been working on a a tutorial to explain how to use tools in Galaxy Australia to assemble large (eukaryotic) genomes. The first version of this tutorial is here: https://doi.org/10.5281/zenodo.5655813. I wanted to show an example that researchers can customise for their own data, as well as adding context around the challenges of genome assembly.
-
Overview of tools in the tutorial workflow
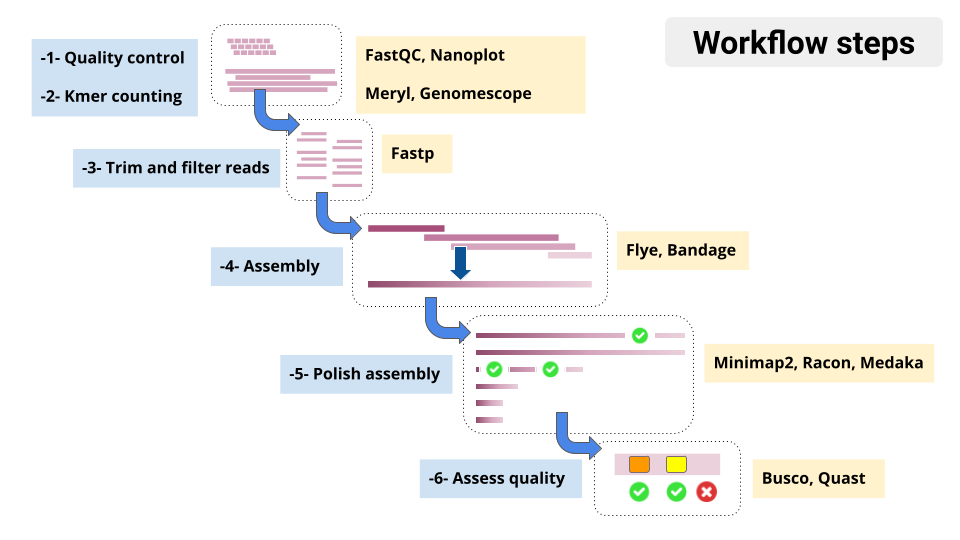
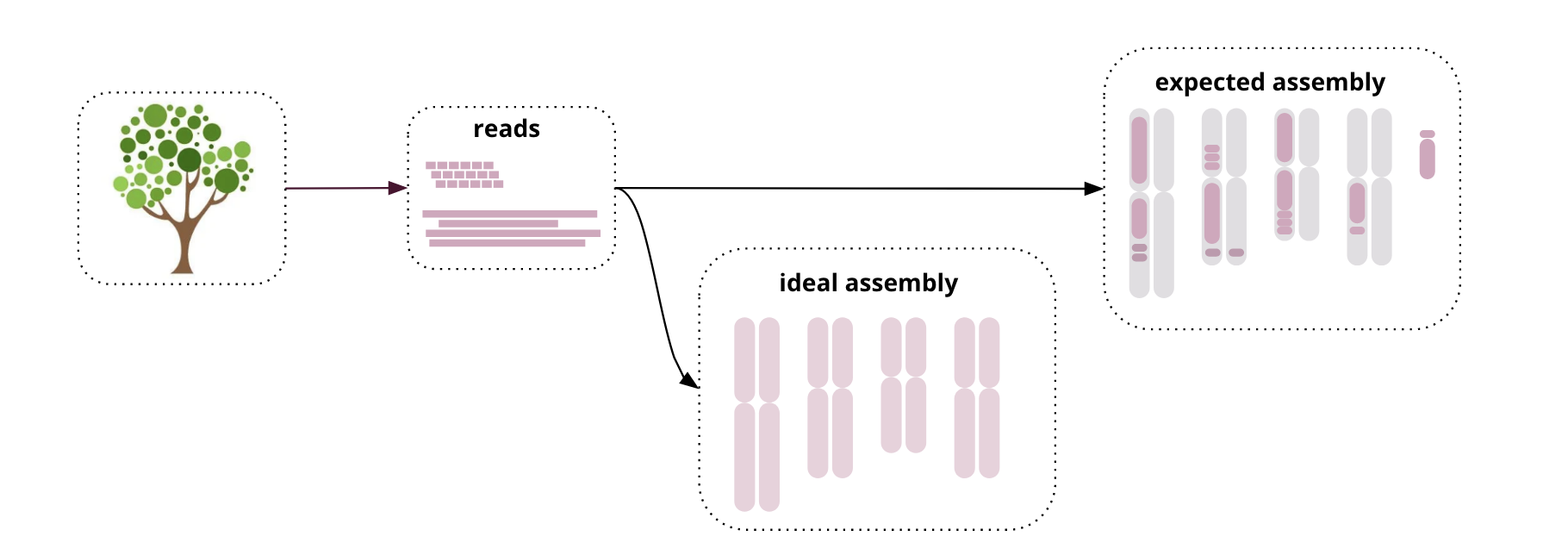
- Genome assembly is hard! We don’t expect perfect, chromosome-sized contigs (yet)

The workflows have been registered on Workflow Hub - see the list here.
August 2021
-
Currently developing workflows and training for large-genome assembly on Galaxy Australia. This project has been summarized in a recent Galaxy conference presentation: Syme A, Christiansen J, Ward N et al. Driving workforce transition in Australian life science research; F1000Research 2021. https://doi.org/10.7490/f1000research.1118591.1.
-
Workflows in development
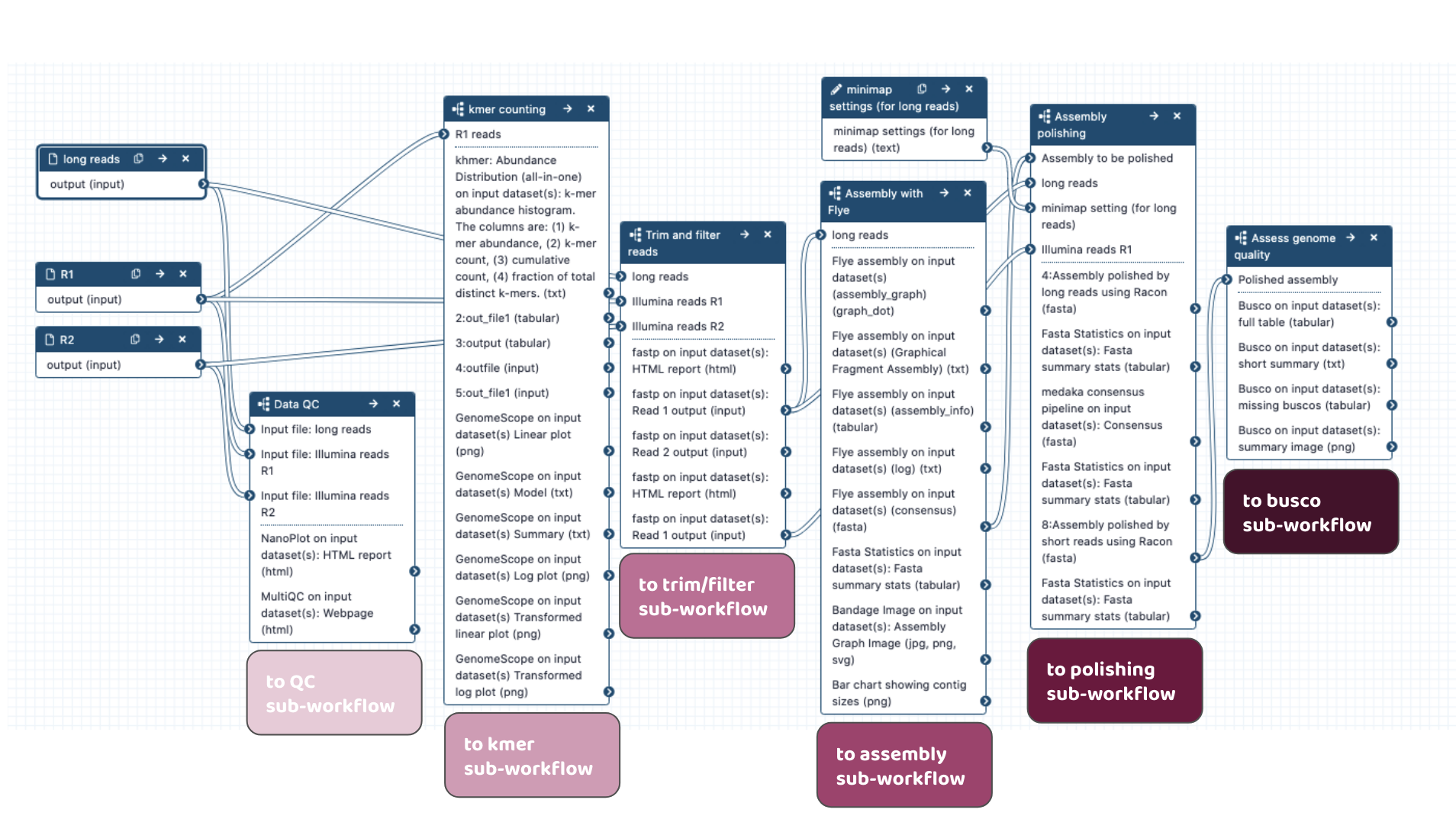
-
It has been great to test some new capacity on Galaxy Australia with the task of assembly a plant genome, with great success (and speed!). Link to report: https://www.biocommons.org.au/news/galaxy-aus-uom-test.
-
Report snippet

April 2021
-
Genome assemblies for the golden wattle (Acacia pycnantha) organelles - the mitochondrial and chloroplast genome. Pre-print: Long-read assemblies reveal structural diversity in genomes of organelles - an example with Acacia pycnantha; Anna E. Syme, Todd G.B. McLay, Frank Udovicic, David J. Cantrill, Daniel J. Murphy; bioRxiv 2020.12.22.423164. https://doi.org/10.1101/2020.12.22.423164
-
One of the most difficult parts of this analysis was to extract the organelle-only reads from the full genomic read sets that contain a mix of nuclear, mitome and plastome reads. One solution is to use iterative baiting (all analysis code here: https://github.com/AnnaSyme/organelle-assembly), shown in this flowchart:
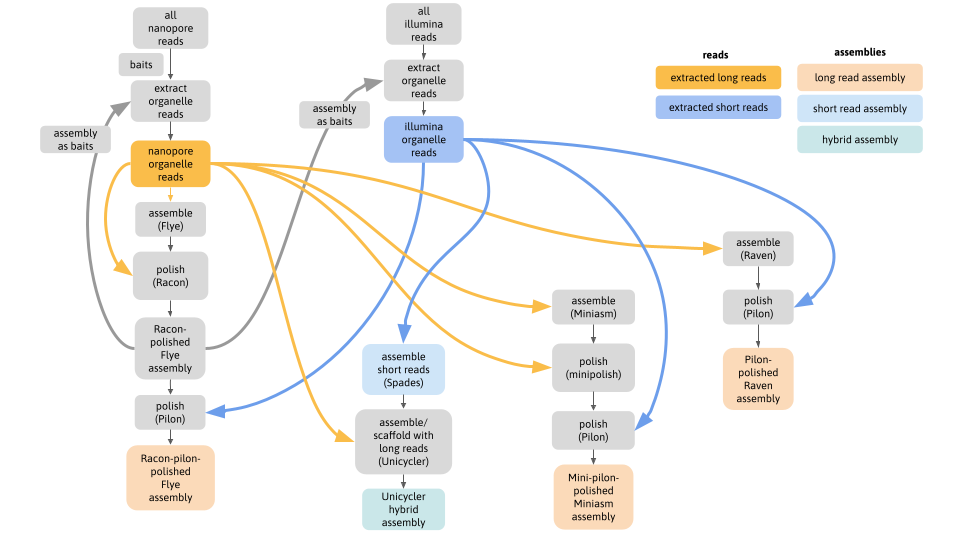
- Although these genomes are often represented as circular, static structures, this work showed that different assemblies revealed contrasting arrangements of genomic segments; a hypothesis supported by mapped reads spanning alternate paths.
The chloroplast genome assembly work has been translated into tutorial materials and used for:
- Galaxy Training Network: Chloroplast genome assembly (Galaxy Training Materials). Syme, Anna. 2021 https://training.galaxyproject.org/training-material/topics/assembly/tutorials/chloroplast-assembly/tutorial.html
- Australian BioCommons webinar (December 2020, https://www.biocommons.org.au/events/chloroplastgenomeassembly)
- The Galaxy Training Network Smörgåsbord (February 2021, https://shiltemann.github.io/global-galaxy-course/)
- Melbourne Bioinformatics Plant Genomics Workshop (April 2021, https://www.melbournebioinformatics.org.au/training-events/plant-genomics-galaxy/)
- The Galaxy Community Conference (July 2021, https://galaxyproject.org/events/gcc2021/training//science-track#introduction-to-ngs-analysis).
October 2020
- A nice welcome here from Melbourne Bioinformatics! “Dr Anna Syme re-joins our team to work on major Australian BioCommons project” https://www.melbournebioinformatics.org.au/news/anna-syme-2020/
Research Scientist, Bioinformatics
Royal Botanic Gardens Victoria, February 2019 to September 2020
I worked on the Genomics for Australian Plants project (Bioplatforms Australia), as lead for the working groups: Bioinformatics (Reference Genomes), and Bioinformatics Training. A focus was the genome for the golden wattle, Acacia pycnantha, with sequencing data from Illumina, Oxford Nanopore, and 10X. A manuscript for assemblies of the chloroplast and mitochondrial genomes is in prep. Long sequencing reads have been particularly informative regarding the structural complexity of the mitome.
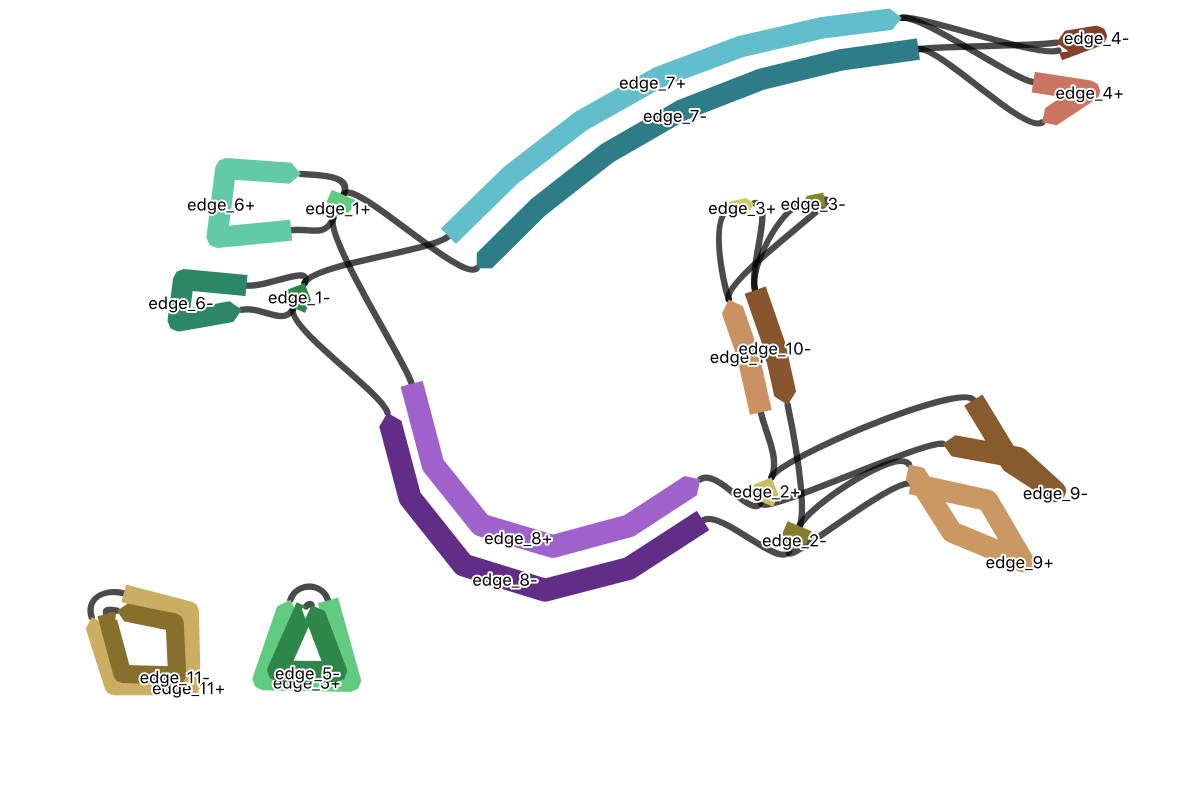
Investigating paths through the mitochondrial assembly to understand genomic structure
The training webpage for the Australian plant genomics research community has been set up and will expand over the next few years.
Snapshot of training webpages, Feb 2020: https://plant-genomics.github.io/training/
New training will be developed using both the Galaxy Australia platform and command-line tools. A recent publication aims to make bioinformatics software development easier and more user-friendly with the Bionitio tool:
Overview of the automated process for creating new projects performed by the Bionitio bootstrap script. From: Peter Georgeson, Anna Syme, Clare Sloggett, Jessica Chung, Harriet Dashnow, Michael Milton, Andrew Lonsdale, David Powell, Torsten Seemann, Bernard Pope. (2019) Bionitio: demonstrating and facilitating best practices for bioinformatics command-line software, GigaScience.
Bioinformatician
Melbourne Bioinformatics, May 2016 to January 2019
Bacterial genome assembly for Bioplatforms Australia project on antibiotic resistance. Development and presentation of bioinformatics training. Galaxy Australia Training Manager (2018). Contributor to the Galaxy Training Network. Subject coordinator for Elements of Bioinformatics (Masters level, University of Melbourne)(2018).
Training examples: Galaxy Australia training webpage snapshot, https://galaxy-au-training.github.io/tutorials/; Development of long-read genome assembly workshop (command-line, presented at Melbourne Bioinformatics).
Molecular Systematist
Royal Botanic Gardens Victoria, September 2010 to December 2012
Molecular analyses of grasses (Stipeae) and fungi (Agaricaceae), including studies on diversification rates, and the evolution of morphological features. Molecular Lab Manager.
From Syme, A. E. (2012). Diversification rates in the Australasian endemic grass Austrostipa - 15 million years of constant evolution. Plant Systematics and Evolution; and Syme, A. E., Murphy, D. J., Holmes, G. D., Gardner, S., Fowler, R. and Cantrill, D. C. (2012). An expanded phylogenetic analysis of Austrostipa (Poaceae: Stipeae) to test infrageneric relationships. Australian Systematic Botany, 25:1-10.
From Lebel, T. and Syme, A. E. (2012). Sequestrate Agaricus and Macrolepiota from Australia: new combinations and species, and their position in a calibrated phylogeny. Mycologia, 104:496-520.
Postdoctoral Researcher
UC Santa Barbara, California, April 2007 to April 2008
At the Oakley Evolution Lab. Evolutionary dynamics of ostracods including modeling gain/loss of the lateral eye and modeling speciation/extinction rates. Phylogenetics of vision genes in Pancrustacea.
From Syme A. E. and Oakley T. H. (2012). Dispersal between shallow and abyssal seas and evolutionary loss and re-gain of ostracod compound eyes in cylindroleberidid ostracods: conflicting conclusions from different comparative methods. Systematic Biology, 61(2): 314-336.
From Lum, K. E., Syme, A. E., Schwab, A. K. and Oakley, T. H. (2008). Euphilomedes chupacabra (Ostracoda: Myodocopida: Philomedidae), a new demersal marine species from coastal Puerto Rico with male-biased vespertine swimming activity. Zootaxa, 1684: 35–57.
From Rivera, A. S, Pankey, M. S., Plachetzki, D. C., Villacorta, C., Syme, A. E., Serb, J. M., Omilian, A. R. and Oakley, T. H. (2010). Gene duplication and the origins of morphological complexity in pancrustacean eyes, a genomic approach. BMC Evolutionary Biology, 10:123.
PhD (Zoology)
The University of Melbourne, Australia, January 2003 - January 2007
Systematics, taxonomy, evolution of the ostracod family Cylindroleberididae (Crustacea). Co-author on marine field guides.
From Syme, A. E. and Poore, G. C. B. (2006). Three new species of the Cylindroleberididae from coastal Australian waters (Crustacea: Ostracoda). Zootaxa, 1305: 51–67.
From Syme and Poore (2008) PLoS ONE, A Supplementary Description of Cypridina mariae and Rediagnosis of the Genus Cylindroleberis (Ostracoda: Myodocopa: Cylindroleberididae) https://doi.org/10.1371/journal.pone.0001960.
Leuroleberis mackenziei and Asteropterygion magnum. Collected Victoria, Australia, 2004.
Wilson, R., Norman, M. and Syme, A. E. (2007). An Introduction to Marine Life. Museum Victoria; CSIRO Publishing. Poore, G. C. B. and Syme, A. E. (2009). Barnacles. Museum Victoria; CSIRO Publishing.











